Drug discovery is a costly and time-intensive process, with binding free energy calculations between the potential drug molecule and the target being crucial for reducing drug discovery costs, streamlining the process and avoiding dead-end leads.
Researchers are left with the challenge of predicting how strongly a potential drug molecule will bind to its target, so there is a high demand for computational approaches that can design compounds that bind effectively to the target molecules. However, traditional methods to calculate free energy binding struggle when faced with large chemical transformations or significant molecular rearrangements.
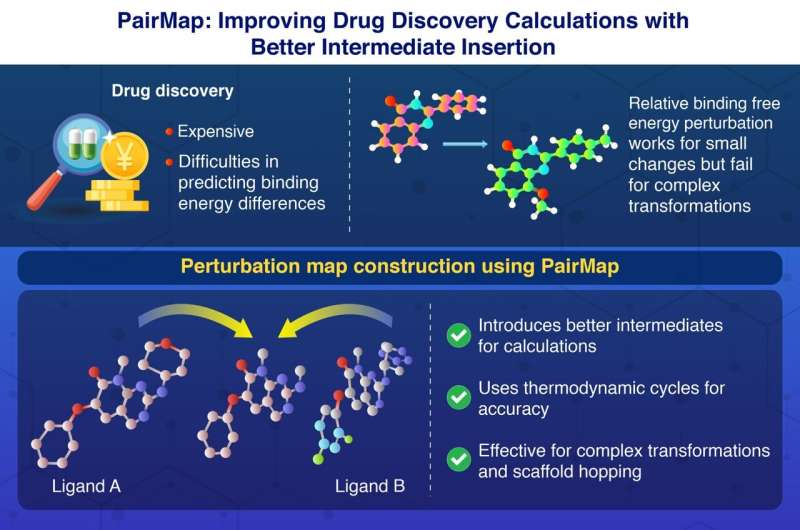
This scenario has led to the development of PairMap, a computational approach developed to address these challenges head-on. Relative binding free energy perturbation (RBFEP) calculations have long been used to predict the binding affinities of chemically similar compounds.
While effective for small structural changes, these calculations falter when tasked with large transformations, leading to high errors and computational inefficiencies. PairMap overcomes this limitation by systematically introducing intermediate compounds to create a step-by-step pathway between the two molecules in question. This novel approach minimizes calculation errors, improves convergence, and reduces computational costs.
This study, a joint effort between Associate Professor Masahito Ohue from Institute of Science Tokyo (Science Tokyo) Japan, and Alivexis, Inc., is a highlight of their close partnership aimed at advancing cutting-edge research. Published online in the Journal of Chemical Information and Modeling on January 12, 2025, the study showcases groundbreaking developments in drug discovery.
“PairMap has the potential to redefine drug discovery. By incorporating well-constructed intermediates and thermodynamic cycles, we’ve achieved an unparalleled level of precision in our energy predictions,” says Ohue.
The team used two ligands (molecules that can bind to metal atoms) named “A” and “B.” The researchers initially generated possible intermediates that can arise in the reaction between ligands A and B. They determined the optimal intermediate pathway that can result in effective binding between A and B and then constructed a perturbation map, which can reveal the binding free energy between these ligands.
In a comprehensive evaluation using benchmark datasets, PairMap showcased its superiority over conventional methods. It reduced the mean absolute error in binding energy predictions from 1.70 kcal/mol (using traditional methods) to 0.93 kcal/mol. For complex transformations, PairMap achieved exceptional accuracy compared to existing approaches, like absolute binding free energy perturbation and RBFEP. The results aligned closely with experimental data, offering a compelling validation of the tool’s practical applications.
What sets PairMap apart is its comprehensive approach to intermediate generation. Unlike traditional methods that use a threshold-based approach to introduce intermediates, PairMap exhaustively explores all potential intermediates and identifies the optimal conversion path. Additionally, it incorporates thermodynamic cycles into its calculations, ensuring high accuracy through error correction based on the conservation of energy.
Ohue highlighted the broader implications of the approach, “By addressing the most complex chemical transformations with minimal computational overhead, PairMap empowers researchers to explore chemical spaces previously considered too challenging.”
Beyond its current achievements, PairMap has exciting implications for the future of computational drug design. It supports the creation of more effective and targeted medications, potentially accelerating the discovery of treatments for diseases that currently lack effective therapies. Moreover, the tool’s flexibility allows for seamless integration into existing drug discovery workflows, reducing costs and time-to-market for new drugs.
Future developments aim to extend its application to cases involving intermediates with significant charge changes. By making this tool accessible, both Professor Ohue at Science Tokyo and his collaborators at Alivexis, Inc. hope to catalyze a new era of innovation in drug discovery.
More information:
Kairi Furui et al, PairMap: An Intermediate Insertion Approach for Improving the Accuracy of Relative Free Energy Perturbation Calculations for Distant Compound Transformations, Journal of Chemical Information and Modeling (2025). DOI: 10.1021/acs.jcim.4c01634
Provided by
Institute of Science Tokyo
Citation:
Precise binding free energy calculations unlock new frontiers in drug design (2025, February 4)
retrieved 4 February 2025
from https://phys.org/news/2025-02-precise-free-energy-frontiers-drug.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.